Note
Go to the end to download the full example code
Single timestep#
Illustrates setting up a simulation and solving at a single time step
from platform import release
import re
import numpy as np
import xcell as xc
import matplotlib.pyplot as plt
Set simulation preferences#
# Misc parameters
study_path = "/dev/null"
# options = uniform, adaptive
meshtype = "adaptive"
max_depth = 10 # Maximum successive splits allowed for octree mesh
nX = 10 # Number of elements along an axis for a uniform mesh
# options: Admittance, Face, FEM
element_type = "Admittance"
dual = True
regularize = False
# options: analytical, ground
boundaryType = "ground"
fixedSource = False # otherwise, simulate current injection
Run simulation#
xmax = 1e-4 # domain boundary
rElec = 1e-6 # center source radius
sigma = np.ones(3)
bbox = np.append(-xmax * np.ones(3), xmax * np.ones(3))
study = xc.Study(study_path, bbox)
setup = study.new_simulation()
setup.mesh.element_type = element_type
setup.meshtype = meshtype
geo = xc.geometry.Sphere(center=np.zeros(3), radius=rElec)
if fixedSource:
setup.add_voltage_source(xc.signals.Signal(1), geo)
srcMag = 1.0
srcType = "Voltage"
else:
srcMag = 4 * np.pi * sigma[0] * rElec
setup.add_current_source(xc.signals.Signal(srcMag), geo)
srcType = "Current"
if meshtype == "uniform":
setup.make_uniform_grid(nX)
print("uniform, %d per axis" % nX)
else:
setup.make_adaptive_grid(
ref_pts=np.zeros((1, 3)),
max_depth=np.array(max_depth, ndmin=1),
min_l0_function=xc.general_metric,
# coefs=np.array(2**(-0.2*max_depth), ndmin=1))
coefs=np.array(0.2, ndmin=1),
)
if boundaryType == "analytical":
boundary_function = None
else:
def boundary_function(coord):
r = np.linalg.norm(coord)
return rElec / (r * np.pi * 4)
setup.finalize_mesh()
setup.set_boundary_nodes(boundary_function, sigma=1)
v = setup.solve()
setup.apply_transforms()
setup.getMemUsage(True)
setup.print_total_time()
setup.start_timing("Estimate error")
# srcMag,srcType,showPlots=showGraphs)
errEst, arErr, _, _, _ = setup.calculate_errors()
print("error: %g" % errEst)
setup.log_time()
bnd = setup.mesh.bbox[[0, 3, 2, 4]]
927.842 Mb used
Total time: 2.15392s [CPU], 1.45783s [Wall]
error: 0.217408
ax = plt.figure().add_subplot()
xc.visualizers.format_xy_axis(ax, bnd)
arr = xc.visualizers.resample_plane(ax, setup)
colormap, color_norm = xc.visualizers.get_cmap(arr.ravel(), forceBipolar=True)
xc.visualizers.patchwork_image(ax, [arr], colormap, color_norm, extent=bnd)
_, _, edge_points = setup.get_elements_in_plane()
xc.visualizers.show_2d_edges(ax, edge_points)
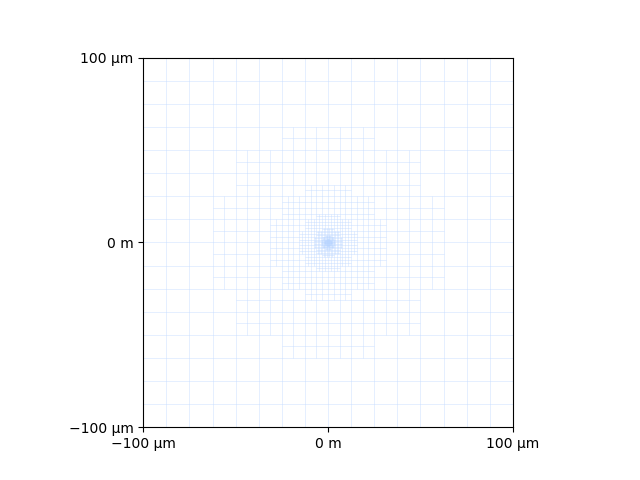
<matplotlib.collections.LineCollection object at 0x2ccf7af10>
TOPOLOGY/connectivity
ax = xc.visualizers.show_mesh(setup)
ax.set_xticks([])
ax.set_yticks([])
ax.set_zticks([])
ghost = (0.0, 0.0, 0.0, 0.0)
ax.xaxis.set_pane_color(ghost)
ax.yaxis.set_pane_color(ghost)
ax.zaxis.set_pane_color(ghost)
xc.visualizers.show_3d_edges(ax, setup.mesh.node_coords, setup.edges, setup.conductances)
bnodes = setup.mesh.get_boundary_nodes()
xc.visualizers.show_3d_nodes(ax, setup.mesh.node_coords[bnodes], node_values=np.ones_like(bnodes), colors="r")
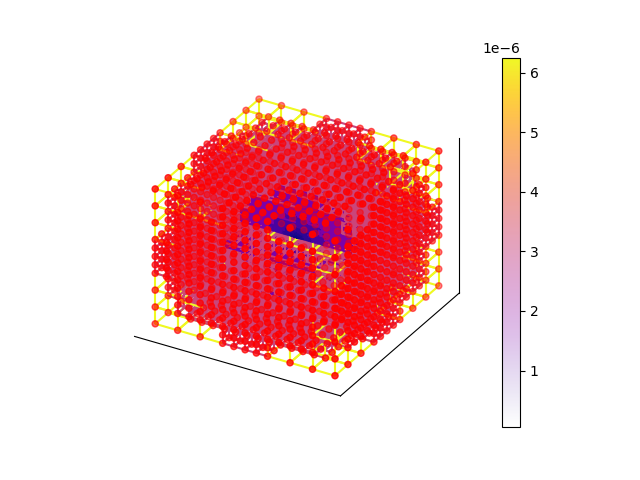
<mpl_toolkits.mplot3d.art3d.Path3DCollection object at 0x2ccfde910>
SliceSet#
# sphinx_gallery_thumbnail_number = 4
img = xc.visualizers.SliceSet(plt.figure(), study)
img.add_simulation_data(setup, append=True)
_ = img.get_artists(0)
![Simulated potential [V], Absolute error [V]](../_images/sphx_glr_plot_singleStep_003.png)
ErrorGraph#
ptr = xc.visualizers.ErrorGraph(plt.figure(), study)
ptr.prefs["universalPts"] = True
pdata = ptr.add_simulation_data(setup)
_ = ptr.get_artists(0, pdata)
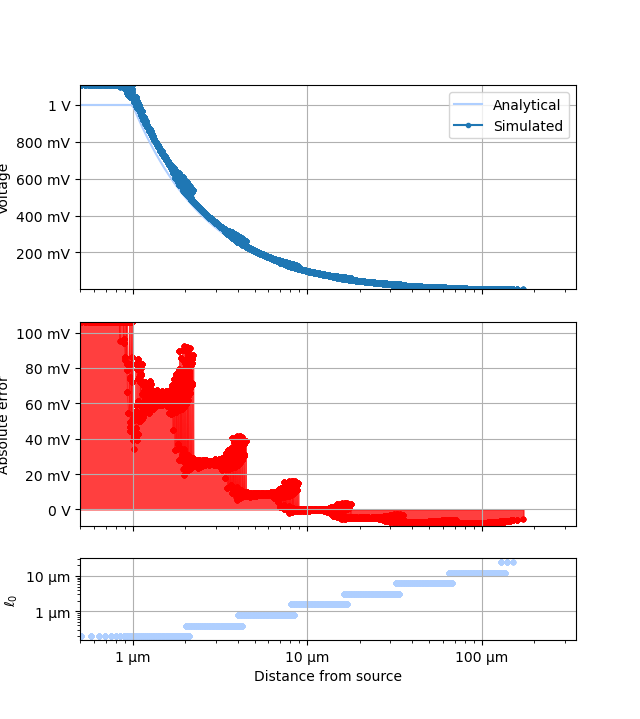
LogError#
P = xc.visualizers.LogError(None, study)
P.add_simulation_data(setup, True)
_ = P.get_artists(0)
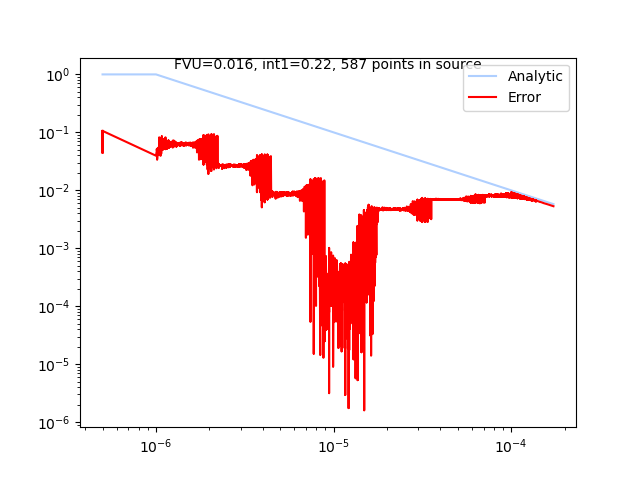
Total running time of the script: (1 minutes 7.762 seconds)