Note
Go to the end to download the full example code
DBS Electrode#
Programmatically generate micro-macro electrode array
import xcell as xc
import numpy as np
from scipy.spatial.transform import Rotation
import pyvista as pv
import pickle
from matplotlib.colors import to_rgba_array
composite = pv.MultiBlock()
regions = xc.io.Regions()
# bodies, misc, oriens, and hilus
sigmas = 1 / np.array([6.429, 3.215, 2.879, 2.605])
# latest from https://github.com/Head-Conductivity/Human-Head-Conductivity
sigma_0 = 0.3841
xdom = 5e-2
dbody = 1.3e-3
dmicro = 17e-6
# dmicro = 5e-4
wmacro = 2e-3
pitchMacro = 5e-3
nmacro = 6 # 4-6
nmicro = 24 # 10-24
microRows = 5
microCols = 3
orientation = np.array([1.0, 0.0, 0.0])
tipPt = 1e-3 * np.array([-5.0, 2.0, 0])
bodyL = 0.05
tpulse = 1e-3 # per phase
ipulse = 150e-6
vpulse = 1.0
hippo = pv.read("./Geometry/slice77600_ext10000.vtk")
brainXform = -np.array(hippo.center)
hippo.translate(brainXform, inplace=True)
files = ["bodies", "misc", "oriens", "hilus"]
for f, sig in zip(files, sigmas):
region = pv.read("Geometry/" + f + ".stl").translate(brainXform)
region.cell_data["sigma"] = sig
regions.addMesh(region, "Conductors")
composite.append(region, name=f)
# xdom = max(np.array(hippo.bounds[1::2])-np.array(hippo.bounds[::2]))
xdom = pitchMacro * (nmacro + 1)
bbox = xdom * np.concatenate((-np.ones(3), np.ones(3)))
# set smallest element to microelectrode radius or smaller
max_depth = int(np.log2(xdom / dmicro)) + 2
sim = xc.Simulation("test", bbox=bbox)
body = xc.geometry.Cylinder(tipPt + bodyL * orientation / 2, radius=dbody / 2,
length=bodyL, axis=orientation)
# bugfix to force detection of points inside cylindrical body
bodyMesh = xc.geometry.to_pyvista(body)
regions.addMesh(bodyMesh, category="Insulators")
# bodyMesh = pv.Cylinder(center=body.center,
# direction=body.axis,
# radius=body.radius,
# height=body.length)
macroElectrodes = []
microElectrodes = []
elecMeshes = []
ref_pts = []
refSizes = []
# Generate macroelectrodes (bands)
for ii in range(nmacro):
pt = tipPt + (ii + 1) * pitchMacro * orientation
geo = xc.geometry.Cylinder(pt, dbody / 2, wmacro, orientation)
sim.add_current_source(xc.signals.Signal(0), geometry=geo)
macroElectrodes.append(geo)
ref_pts.append(geo.center)
regions.addMesh(xc.geometry.to_pyvista(geo), category="Electrodes")
# Generate microelectrodes
for ii in range(microRows):
rowpt = tipPt + (ii + 0.5) * pitchMacro * orientation
for jj in range(microCols):
rot = Rotation.from_rotvec(orientation * 2 * jj / microCols * np.pi)
microOrientation = rot.apply(0.5 * dbody * np.array([0.0, 0.0, 1.0]))
geo = xc.geometry.Disk(center=rowpt + microOrientation,
radius=dmicro / 2, axis=microOrientation,
tol=0.5)
sim.add_current_source(xc.signals.Signal(0), geometry=geo)
microElectrodes.append(geo)
ref_pts.append(geo.center)
regions.addMesh(xc.geometry.to_pyvista(geo), category="Electrodes")
p = xc.visualizers.PVScene()
p.setup(regions, opacity=0.5)
p.show()
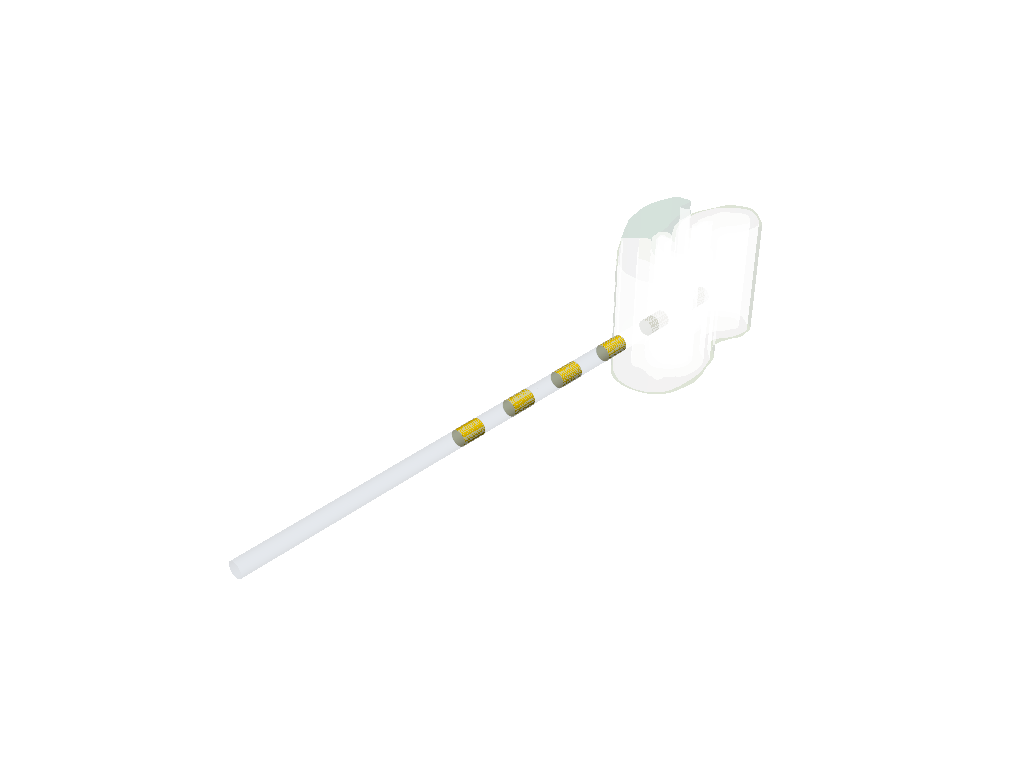
sim.quick_adaptive_grid(max_depth)
vmesh = xc.io.to_vtk(sim.mesh)
vmesh.cell_data["sigma"] = sigma_0
regions.assign_sigma(sim.mesh, default_sigma=sigma_0)
sim.current_sources[nmacro + 1].value = 150e-6
sim.set_boundary_nodes()
v = sim.solve()
vmesh.point_data["voltage"] = v
vmesh.set_active_scalars("voltage")
(<FieldAssociation.POINT: 0>, pyvista_ndarray([0., 0., 0., ..., 0., 0., 0.]))
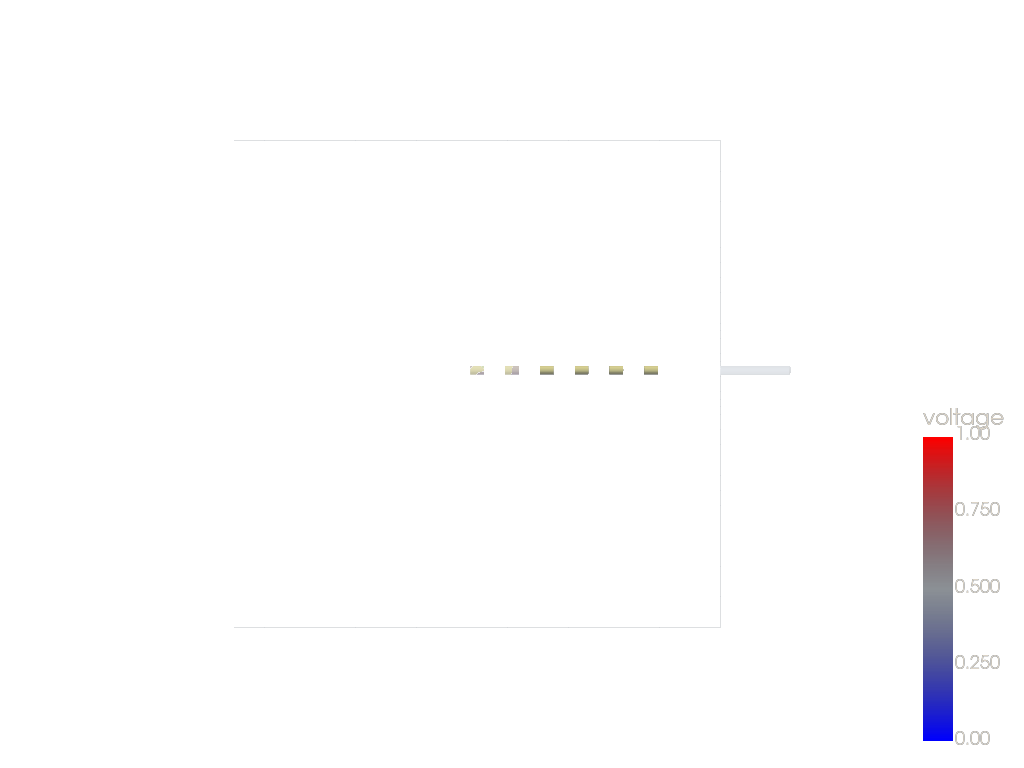
p = xc.visualizers.PVScene()
p.setup(regions) # , mesh=vmesh, simData='voltage')
# p.camera.tight(padding=0.1)
p.add_mesh(vmesh.slice(normal="z"), show_edges=True, cmap=xc.colors.CM_BIPOLAR)
cambox = np.array(hippo.bounds)
cambox[4:] = 0.0
# p.reset_camera(bounds=cambox)
p.view_xy()
p.show()
# sphinx_gallery_thumbnail_number = 3
# Save outputs
regions.save("Geometry/composite.vtm")
Total running time of the script: (0 minutes 51.256 seconds)